×
Network disruptions
We have been experiencing disruptions on our local network which has affected the stability of these web pages.
We have been working with IT support team to get this fixed as a matter of urgency and apologise for any inconvenience.
PDB Information
| PDB | 5C1E |
| Method | X-RAY DIFFRACTION |
| Host Organism | Saccharomyces cerevisiae |
| Gene Source | Aspergillus niger (strain ATCC 1015 / CBS 113.46 / FGSC A1144 / LSHB Ac4 / NCTC 3858a / NRRL 328 / USDA 3528.7) |
| Primary Citation |
Structure and Properties of a Non-processive, Salt-requiring, and Acidophilic Pectin Methylesterase from Aspergillus niger Provide Insights into the Key Determinants of Processivity Control.
Kent, L.M., Loo, T.S., Melton, L.D., Mercadante, D., Williams, M.A., Jameson, G.B.
J.Biol.Chem.
|
| Header | Hydrolase |
| Released | 2015-06-13 |
| Resolution | 1.750 |
| CATH Insert Date | 16 Aug, 2015 |
PDB Images (3)
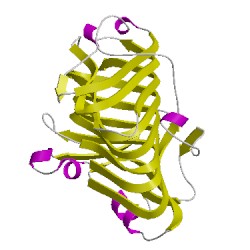
5c1eA00
CATH Domain 5c1eA00
PDB Chains (1)
| Chain ID | Date inserted into CATH | CATH Status |
| A | 17 Aug, 2015 | Chopped
|
CATH Domains (1)
UniProtKB Entries (1)
| Accession |
Gene ID |
Taxon |
Description |
| G3YAL0 |
G3YAL0_ASPNA |
Aspergillus niger ATCC 1015 |
Pectinesterase |
